Create a beautifully simple website or blog in under 10 minutes.
** The Dries lab will open its doors on August 1st, see www.drieslab.com !**
I am a computational biologist currently working at Dana-Farber Cancer Institute as a post-doc in the labs of Rani George, Guo-Cheng Yuan and Kwok-Kin Wong (untill the lab moved to NYU). I was born in Belgium where I did my Bachelor and Master in Biomedical Sciences at KU Leuven and was offered a scholarship to perform my master thesis in developmental biology in the lab of Anming Meng at Tsinghua University in China. This thesis was a close collaboration with the group of Danny Huylebroeck at KU Leuven, where I did my PhD studies and started a project at the crossroads of development and systems biology.
During my PhD I became interested in transcriptional dynamics, single-cells and genomics and gradually moved away from the (wet) bench to a (dry) desk. As a post-doc I continued in this direction and I am specifically interested in how individual cells from a multi-cellular organism react to changes in their environment. What is the immediate transcriptional respons and its implication? How do they reach or (un-)lock their current transcriptional state? What is the contribution of cellular heterogeneity and the spatial microenvironment, such as a specialized niche or the immune system? I started to embark on some of these questions in drug treatment and/or resistance models for small-cell lung cancer and neuroblastoma and I’m looking forward to expand our acquired insights in those and other types of cancer and model organisms.
Alltogether, I am a biology explorer with extensive wet and dry lab training (both 5+ years), which enables me to facilitate the interaction between highly-specialized fields. In this role I’ve led and contributed in conceptualizing and driving complex projects, that required high-dimensional analysis, which ultimately lead to a better biological understanding. Altogether, I believe that research is - more than ever - a teamsport which requires a collaborative effort to fully understand and utilize the generated data to answer biological questions. As such, I believe that groundbreaking science is based on 3 fundamental pillars, (1) well-crafted and thoughtfull experiments that embrace (2) novel technologies and are led by (3) data-driven decisions.
Research Fellow in computational biology, 2016 - current
DFCI & HSPH, Boston USA
PhD in Biomedical Sciences, 2017
University of Leuven, Belgium
PhD in Medical Sciences, 2016
Rotterdam Erasmus University, the Netherlands
Master in Biomedical sciences, 2009
University of Leuven, Belgium
BSc in Biomedical sciences, 2007
University of Leuven, Belgium

Create a beautifully simple website or blog in under 10 minutes.
Here you can download my CV
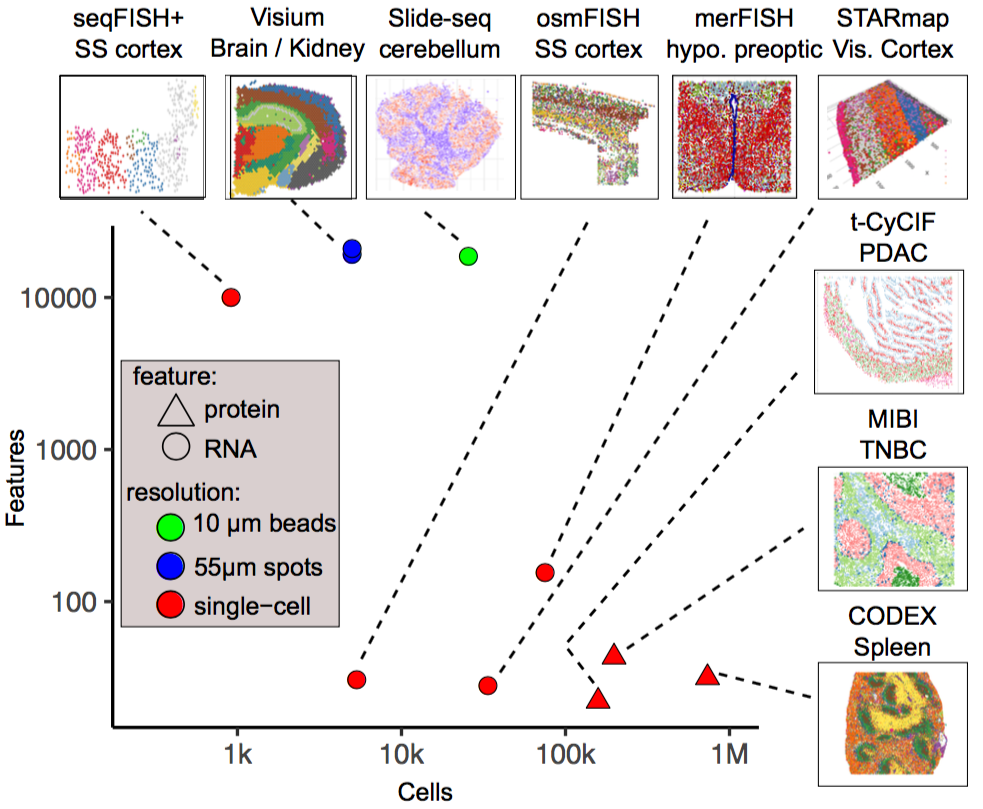
Exploration of the spatial cellular microenvironment. What are the sources of spatial variability? How do cells communicate? How can we integrate different spatial and single-cell technologies?
Cancer genomics
Immunology in cancer
Exploration of single-cell genomics
Stem cells and embryonic development
Transcriptional regulation